Canine transmissible venereal tumors (CTVT) spread when cancer cells move from one dog to the other during sexual contact, without the help of a virus or bacterium. Credit: Ernesto del Aguila III, NHGRI
National Human Genome Research Institute (NHGRI) (CC BY 2.0)
By Karen Graham
A contagious canine cancer that conquered the world by spreading between dogs during mating likely arose around 6,000 years ago in Asia and spread around the globe through maritime activities, scientists say.
A detailed genetic study, published in the journal Science, reveals some surprising, and even mysterious, findings about canine transmissible venereal tumor — a cancer that has survived for thousands of years, having mutated and evolved over time.
This fascinating cancer spreads between dogs through the transfer of living cancer cells, primarily during mating, resulting in genital tumors in both male and female domestic dogs. But what is remarkable about this tumor is that the cancer cells are those of the original dog in which the cancer arose.
The only difference in the modern cancer cells and the cells of the original dog are those that have arisen over time either through spontaneous changes in the cells’ DNA or through changes caused by carcinogens.
Ph.D. student, Adrian Baez-Ortega, one of the authors of the study said, “This tumor has spread to almost every continent, evolving as it spreads…changes to its DNA tell a story of where it has been and when, almost like a historical travel journal.”
The study was led by researchers with the Transmissible Cancer Group at the University of Cambridge. The research team studied tumors taken from 546 canines worldwide – comparing the DNA. They created a phylogenetic tree – a type of family tree of the different mutations in the tumors.
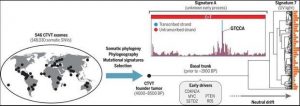
Cancer evolution over thousands of years.
The evolutionary process results in constant change – and means that cells at opposite ends of a tumor can be genetically different. By comparing the patterns of mutations and working backward, scientists can identify which mutations arose when and where, and work out how a tumor originated, evolved, and spread.
While this type of examination usually is done from samples taken from a single body, the researchers used samples collected from six continents. “We mapped out a metastasis on a global scale, over thousands of years,” said Elizabeth Murchison of the University of Cambridge, who led the study.
Based on their genetic studies, the research team concluded that all the tumors found in North, Central and South America came from the same origin point from the same introduction event. After being transferred to the Americas about 500 years ago, it is believed that the disease was then spread to Africa and back to India, according to SciTech Europa.
While the finding shed light on the historical spread of the disease, it is the tumor’s evolution that particularly excites the researchers. With recent advances in cancer research, scientists are able to look closer at mutations in tumor cells and identify unique signatures left by carcinogens. This allows them to see, for example, the damage that ultraviolet (UV) light causes.
Using these new techniques, researchers were able to identify the signatures for five different biological processes that have damaged the canine tumor over its history, including exposure to UV light. A new signature – called “Signature A” – has a very distinctive mutational signature, and has never been seen before in a tumor. It caused mutations only in the tumor’s distant past, several thousand years ago, and has never been seen since.
“This is really exciting – we’ve never seen anything like the pattern caused by this carcinogen before,” says Dr. Murchison. “It looks like the tumor was exposed to something thousands of years ago that caused changes to its DNA for some length of time and then disappeared. It’s a mystery what the carcinogen could be. Perhaps it was something present in the environment when the cancer first arose.”
From Digital Journal
Somatic evolution and global expansion of an ancient transmissible cancer lineage
Science 02 Aug 2019:
Vol. 365, Issue 6452, eaau9923
DOI: 10.1126/science.aau9923
Structured Abstract
INTRODUCTION
The canine transmissible venereal tumor (CTVT) is a sexually transmitted cancer that manifests as genital tumors in dogs. This cancer first arose in an individual “founder dog” several thousand years ago and has since survived by transfer of living cancer cells to new hosts during coitus. Today, CTVT affects dogs around the world and is the oldest and most prolific known cancer lineage. CTVT thus provides an opportunity to explore the evolution of cancer over the long term and to track the unusual biological transition from multicellular organism to obligate conspecific asexual parasite. Furthermore, the CTVT genome, acting as a living biomarker, has recorded the changing mutagenic environments experienced by this cancer throughout millennia and across continents.
RATIONALE
To capture the genetic diversity of the CTVT lineage, we analyzed somatic mutations extracted from the protein-coding genomes (exomes) of 546 globally distributed CTVT tumors. We inferred a time-resolved phylogenetic tree for the clone and used this to trace the worldwide spread of the disease and to select subsets of mutations acquired at known geographical locations and time periods. Computational methods were applied to extract mutational signatures and to measure their exposures across time and space. In addition, we assessed the activity of selection using ratios of nonsynonymous and synonymous variants.
RESULTS
The CTVT phylogeny reveals that the lineage first arose from its founder dog 4000 to 8500 years ago, likely in Asia, with the most recent common ancestor of modern globally distributed tumors occurring ~1900 years ago. CTVT underwent a rapid global expansion within the past 500 years, likely aided by intensification of human maritime travel. We identify a highly specific mutational signature dominated by C>T mutations at GTCCA pentanucleotide contexts, which operated in CTVT up until ~1000 years ago. The number of mutations caused by ultraviolet light exposure is correlated with latitude of tumor collection, and we identify CTVTs with heritable hyperactivity of an endogenous mutational process. Several “driver” mutation candidates are identified in the basal trunk of the CTVT tree, but there is little evidence for ongoing positive selection. Although negative selection is detectable, its effect is largely confined to genes with known essential functions, thus implying that CTVT predominantly evolves through neutral processes.
CONCLUSION
We have traced the evolution of a transmissible cancer over several thousand years, tracking its spread across continents and contrasting the mutational processes and selective forces that molded its genome with those described in human cancers. The identification of a highly context-specific mutational process that operated in the past but subsequently vanished, as well as correlation of ultraviolet light–induced DNA damage with latitude, highlight the potential for long-lived, widespread clonal organisms to act as biomarkers for mutagenic exposures. Our results suggest that neutral genetic drift is the dominant evolutionary force operating on cancer over the long term, in contrast to the ongoing positive selection that is often observed in short-lived human cancers. The weakness of negative selection in this asexual lineage may be expected to lead to the progressive accumulation of deleterious mutations, invoking Muller’s ratchet and raising the possibility that CTVT may be declining in fitness despite its global success.
Cancer evolution over thousands of years.
The canine transmissible venereal tumor (CTVT) is an ancient contagious cancer with a global distribution. We sequenced the exomes of 546 CTVT tumors and identified somatic single-nucleotide variants (SNVs). These were used to construct a time-resolved phylogenetic tree, yielding insights into the cancer’s phylogeography, mutational processes, and signatures of selection across thousands of years. Notably, a highly context-specific mutational pattern named signature A was identified, which was active in the past but ceased to operate about 1000 years ago. BP, years before present.